User:MirankerAD/RNA Tertiary Structure
THIS PAGE IS NO LONGER ACTIVE. DO NOT EDIT HERE, GO INSTEAD TO RNA Tertiary Structure
[edit]
RNA molecules are capable of diverse functions ranging from molecular recognition to catalysis. Such functions require a precise three dimensional structure. While such structures are diverse and seemingly complex, they are comprised of recurring, easily recognizable tertiary structure motifs that serve as molecular building blocks. Some of the most common motifs for RNA tertiary structure formation are described below, but it is important to remember that this information is based on a limited number of solved RNA structures. Many more RNA tertiary structural motifs will be revealed as new RNA molecules are structurally characterized.
Motifs
[edit]Coaxial stacking
[edit]
Coaxial stacking, otherwise known as helical stacking, is a major determinant of higher order RNA tertiary structure. Coaxial stacking occurs when two RNA duplexes form a contiguous helix, which is stabilized by base stacking at the interface of the two helices. Coaxial stacking was noted in the crystal structure of tRNAPhe[2]. More recently, coaxial stacking has been observed in higher order structures of many ribozymes, including many forms of the self-splicing group I and group II introns. Common coaxial stacking motifs include the kissing loop interaction and the pseudoknot. The stability of these interactions can be predicted by an adaptation of “Turner’s rules”.
In 1994, Walter and Turner determined the free energy contributions of nearest neighbor stacking interactions within a helix-helix interface by using a model system that created a helix-helix interface between a short oligomer and a four-nucleotide overhang at the end of a hairpin stem . Their experiments confirmed that the thermodynamic contribution of base-stacking between two helical secondary structures closely mimics the thermodynamics of standard duplex formation (nearest neighbor interactions predict the thermodynamic stability of the resulting helix). The relative stability of nearest neighbor interactions can be used to predict favorable coaxial stacking based on known secondary structure. Walter and Turner found that, on average, prediction of RNA structure improved from 67% to 74% accuracy when coaxial stacking contributions were included[3]. Theories of coaxial stacking can be tested using the technique of helical fusion. This approach was used by Murphy and Cech to confirm a coaxial stacking interaction between the P4 and P6 helices within the catalytic center of the Tetrahymena group I intron.
Most well-studied RNA tertiary structures contain examples of coaxial stacking. Some prominent examples are tRNA-Phe, group I introns, group II introns, and ribosomal RNAs. Crystal structures of tRNA revealed the presence of two extended helices that result from coaxial stacking of the amino-acid acceptor stem with the T-arm, and stacking of the D- and anticodon-arms. These interactions within tRNA orient the anticodon stem perpendicularly to the amino-acid stem, leading to the functional L-shaped tertiary structure[4]. In group I introns, the P4 and P6 helices were shown to coaxially stack using a combination of biochemical [5] and crystallographic methods. The P456 crystal structure provided a detailed view of how coaxial stacking stabilizes the packing of RNA helices into tertiary structures [6]. In the self-splicing group II intron from Oceanobacillus iheyensis, the IA and IB stems coaxially stack and contribute to the relative orientation of the constituent helices of a five-way junction [7]. This orientation facilitates proper folding of the active site of the functional ribozyme. The ribosome contains numerous examples of coaxial stacking, including stacked segments as long as 70 bp [8].
Two common motifs involving coaxial stacking are kissing loops and pseudoknots. In kissing loop interactions, the single-stranded loop regions of two hairpins interact through base pairing, forming a composite, coaxially stacked helix. Notably, this structure allows all of the nucleotides in each loop to participate in base-pairing and stacking interactions. This motif was visualized and studied using NMR analysis by Lee and Crothers[9]. The pseudoknot motif occurs when a single stranded region of a hairpin loop basepairs with an upstream or downstream sequence within the same RNA strand. The two resulting duplex regions often stack upon one another, forming a stable coaxially stacked composite helix. One example of a pseudoknot motif is the highly stable Hepatitis Delta virus ribozyme, in which the backbone shows an overall double pseudoknot topology[10].
Role of metal ions
[edit]Functional RNAs are often folded, stable molecules with three-dimensional shapes rather than floppy, linear strands[11]. Cations are essential for thermodynamic stabilization of RNA tertiary structures. Metal cations that bind RNA can be monovalent, divalent or trivalent. Potassium (K+) is a common monovalent ion that binds RNA. A common divalent ion that binds RNA is magnesium (Mg2+). Other ions including sodium (Na+), calcium (Ca2+) and manganese (Mn2+) have been found to bind RNA in vivo and in vitro. Multivalent organic cations such as spermidine or spermine are also found in cells and these make important contributions to RNA folding. Trivalent ions such as cobalt hexamine or lanthanide ions such as terbium (Tb3+) are useful experimental tools for studying metal binding to RNA [12].
A metal ion can interact with RNA in multiple ways. An ion can associate diffusely with the RNA backbone, shielding otherwise unfavorable electrostatic interactions. This charge screening is often fulfilled by monovalent ions. Site-bound ions stabilize specific elements of RNA tertiary structure. Site-bound interactions can be further subdivided into two categories depending on whether water mediates the metal binding. “Outer sphere” interactions are mediated by water molecules that surround the metal ion. For example, magnesium hexahydrate interacts with and stabilizes specific RNA tertiary structure motifs via interactions with guanosine in the major groove. Conversely, “inner sphere” interactions are directly mediated by the metal ion. RNA often folds in multiple stages and these steps can be stabilized by different types of cations. In the early stages, RNA forms secondary structures stabilized through the binding of monovalent cations, divalent cations and polyanionic amines in order to neutralize the polyanionic backbone. The later stages of this process involve the formation of RNA tertiary structure, which is stabilized almost largely through the binding of divalent ions such as magnesium with possible contributions from potassium binding.
Metal-binding sites are often localized in the deep and narrow major groove of the RNA duplex, coordinating to the Hoogsteen edges of purines. In particular, metal cations stabilize sites of backbone twisting where tight packing of phosphates results in a region of dense negative charge. There are several metal ion-binding motifs in RNA duplexes that have been identified in crystal structures. For instance, in the P4-P6 domain of the Tetrahymena thermophila group I intron, several ion-binding sites consist of tandem G-U wobble pairs and tandem G-A mismatches, in which divalent cations interact with the Hoogsteen edge of guanosine via O6 and N7 [13][14][15]. Another ion-binding motif in the Tetrahymena group I intron is the A-A platform motif, in which consecutive adenosines in the same strand of RNA form a non-canonical pseudobase pair[16]. Unlike the tandem G-U motif, the A-A platform motif binds preferentially to monovalent cations. In many of these motifs, absence of the monovalent or divalent cations results in either greater flexibility or loss of tertiary structure.
Tetraloop-receptor interactions
[edit]
Tetraloop-receptor interactions combine base-pairing and stacking interactions between the loop nucleotides of a tetraloop motif and a receptor motif located within an RNA duplex, creating a tertiary contact that stabilizes the global tertiary fold of an RNA molecule.
Stem-loops can vary greatly in size and sequence, but tetraloops of four nucleotides are very common and they usually belong to one of three categories, based on sequence [17]. These three families are the CUYG, UNCG and GNRA (see figure on the right) tetraloops. [18]. In each of these tetraloop families, the second and third nucleotides form a turn in the RNA strand and a base-pair between the first and fourth nucleotides stabilizes the stemloop structure. It has been determined, in general, that the stability of the tetraloop depends on the composition of bases within the loop and on the composition of this "closing base pair"[19]. The GNRA family of tetraloops is the most commonly observed within Tetraloop-receptor interactions.

“Tetraloop receptor motifs” are long-range tertiary interactions[20] comprised of hydrogen bonding between the bases in the tetraloop to stemloop sequences in distal sections of the secondary RNA structure [21]. In addition to hydrogen bonding, stacking interactions are an important component of these tertiary interactions. For example, in GNRA-tetraloop interactions, the second nucleotide of the tetraloop stacks directly on an A-platform motif (see above) within the receptor [22]. The sequence of the tetraloop and its receptor often covary so that the same type of tertiary contact can be made with different isoforms of the tetraloop and its cognate receptor [23].
For example, the self-splicing group I intron relies on tetraloop receptor motifs for its structure and function[24] [25]. Specifically, the three adenine residues of the canonical GAAA motif stack on top of the receptor helix and form multiple stabilizing hydrogen bonds with the receptor. The first adenine of the GAAA sequence forms a triple base-pair with the receptor AU bases. The second adenine is stabilized by hydrogen bonds with the same uridine, as well as via its 2'-OH with the recptor and via interactions with the guanine of the GAAA tetraloop. The third adenine forms a triple base pair.
A-minor motif
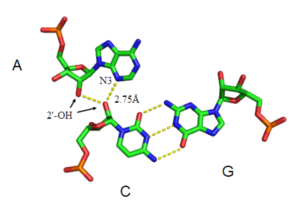
[edit]The A-minor motif is a ubiquitous RNA tertiary structural motif. It is formed by the insertion of an unpaired nucleoside into the minor groove of an RNA duplex. As such it is an example of a minor groove triple. Although guanosine, cytosine and uridine can also form minor groove triple interactions, minor groove interactions by adenine are very common. In the case of adenine, the N1-C2-N3 edge of the inserting base forms hydrogen bonds with one or both of the 2’-OH’s of the duplex, as well as the bases of the duplex (see figure: A-minor interactions). The host duplex is often a G-C basepair.
A-minor motifs have been separated into four classes[26], types 0 to III, based upon the position of the inserting base relative to the two 2’-OH’s of the Watson-Crick base pair. In type I and II A-minor motifs, N3 of adenine is inserted deeply within the minor groove of the duplex (see figure: A minor interactions - type II interaction), and there is good shape complementarity with the base pair. Unlike types 0 and III, type I and II interactions are specific for adenine due to hydrogen bonding interactions. In the type III interaction, both the O2' and N3 of the inserting base are associated less closely with the minor groove of the duplex. Type 0 and III motifs are weaker and non-specific because they are mediated by interactions with a single 2’-OH (see figure: A-minor Interactions - type 0 and type III interactions).
The A-minor motif is among the most common RNA structural motifs in the ribosome, where it contributes to the binding of tRNA to the 23S subunit [27]. They most often stabilize RNA duplex interactions in loops and helices, such as in the core of group II introns[28].
An interesting example of A-minor is its role in anticodon recognition. The ribosome must discriminate between correct and incorrect codon-anticodon pairs. It does so, in part, through the insertion of adenine bases into the minor groove. Incorrect codon-anticodon pairs will present distorted helical geometry, which will prevent the A-minor interaction from stabilizing the binding, and increase the dissociation rate of the incorrect tRNA. [29]
Ribose Zipper
[edit]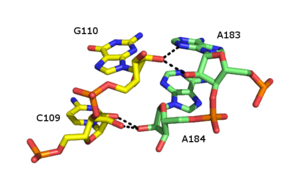
The ribose zipper is an RNA tertiary structural element in which two RNA chains are held together by hydrogen bonding interactions involving the 2’OH of ribose sugars on different strands. The 2'OH can behave as both hydrogen bond donor and acceptor, which allows formation of bifurcated hydrogen bonds with another 2’ OH. [30] Numerous forms of ribose zipper have been reported, but a common type involves four hydrogen bonds between 2'-OH groups of two adjacent sugars. Ribose zippers commonly occur in arrays that stabilize interactions between separate RNA strands [31]. Ribose zippers are often observed as Stem-loop interactions with very low sequence specificity. However, in the small and large ribosomal subunits, there exists a propensity for ribose zippers of the CC/AA sequence- two cytosines on the first chain paired to two adenines on the second chain.
Triplexes and quadruplexes
[edit]Major and Minor Groove Triplexes
The minor groove triple is a ubiquitous RNA structural motif. Because interactions with the minor groove are often mediated by the 2'-OH of the ribose sugar, this RNA motif looks very different from its DNA equivalent. The most common example of a minor loop triple is the A-minor motif, or the insertion of adenosine bases into the minor groove (see above). However, this motif is not restricted to adenosines, as other nucleobases have also been observed to interact with the RNA minor groove.
The minor groove presents a near-perfect complement for an inserted base. This allows for optimal van der Waals contacts, extensive hydrogen bonding and hydrophobic surface burial, and creates a highly energetically favorable interaction[32]. Because minor groove triples are capable of stably packing a free loop and helix, they are key elements in the structure of large ribonucleotides, including the group I intron[33], the group II intron[34], and the ribosome.
Although the major groove of standard A-form RNA is fairly narrow and therefore less available for triplex interaction than the minor groove, major groove triplex interactions can be observed in several RNA structures. These structures consist several combinations of base pair and Hoogsteen interactions. For example, the GGC triplex (GGC amino(N-2)-N-7, imino-carbonyl, carbonyl-amino(N-4); Watson-Crick) observed in the 50S ribosome, composed of a Watson-Crick type G-C pair and an incoming G which forms a pseudo-Hoogsteen network of hydrogen bonding interactions between both bases involved in the canonical pairing [35]. Other notable examples of major groove triplexes include (i) the catalytic core of the group II intron shown in the figure at left [36] (ii) a catalytically essential triple helix observed in human telomerase RNA [37] and (iii) the SAM-II riboswitch. [38]
Quadruplexes
Besides double helices and the above-mentioned triplexes, RNA can also form quadruple helices. There are diverse structures of RNA base quadruplexes. Four consecutive guanine residues can form a quadruplex in RNA by Hoogsteen hydrogen bonds to form a “Hoogsteen ring” (See Figure) [39]. G-C and A-U pairs can also form base quadruplex with a combination of Watson-Crick pairing and noncanonical pairing in the minor groove [40]. The core of malachite green aptamer is also a kind of base quadruplex with a different hydrogen bonding pattern (See Figure) [41] The quadruplex can repeat several times consecutively, producing an immensely stable structure.
The unique structure of quadruplex regions in RNA may serve different functions in a biological system. Two important functions are the binding potential with ligands or proteins, and its ability to stabilize the whole tertiary structure of DNA or RNA. The strong structure can inhibit or modulate transcription and replication, such as in the telomeres of chromosomes and the UTR of mRNA[42]. The base identity is important towards ligand binding. The G-quartet typically binds monovalent cations such as potassium, while other bases can bind numerous other ligands such as hypoxanthine in a U-U-C-U quadruplex[43].
Along with these functions, the G-quadruplex in the mRNA around the ribosome binding regions could serve as a regulator of gene expression in bacteria[44]. There may be more interesting structures and functions yet to be discovered in vivo.
See also
[edit]Stem-loop
Pseudoknot
Secondary_structure_prediction
Group_I_catalytic_intron
Wobble_base_pair
Riboswitch
Group_II_intron
Ribozyme
RNA_structure_probing
Hammerhead_ribozyme
RNA_structure
TRNA
Hoogsteen_base_pair
G-quadruplex
Tetraloop
Ribosome
Rna
Base_pair
DNA
Notes
[edit]The first draft of this page was prepared as an assignment given to Yale University students studying MB&B 420a/720a : Macromolecular Structure and Biophysical Analysis. It was first posted to article space on December 23, 2009.
References
[edit]- Tamura, M., Holbrook, S. "Sequence and Structural Conservation in RNA Ribose Zippers." Journal of Molecular Biology. (2002), 320, 455.
- Batey, R.T., Rambo, R.P., Doudna, J.A. "Tertiary Motifs in RNA Structure and Folding." Angewandte Chemie International Edition. (1999) 38, 2326.
- ^ Toor N., Keating, K.S., Fedorova, O., Rajashankar, K., Wang, J., Pyle, A.M. Tertiary architecture of the Oceanobacillus iheyensis group II intron. RNA (2010), 16, 57-69
- ^ Quigley, GJ. and Rich, A. Structural domains of transfer RNA molecules. Science (1976) 194, 796
- ^ Walter, AE, Turner, DH, Kim, J, Lyttle, MH, Muller, P, Mathews, DH, Zuker, M. Coaxial stacking of helixes enhances binding of oligoribonucleotides and improves predictions of RNA folding. Proc Natl Acad Sci USA, (1994) 91, 9218-9222.
- ^ Quigley, GJ. and Rich, A. Structural domains of transfer RNA molecules. Science (1976) 194, 796
- ^ Murphy et al. (1994) Coaxially Stacked RNA Helices in the Catalytic Center of the Tetrahymena Ribozyme. Science, 265: 1709
- ^ Cate et al. (1996) Crystal Structure of a Group I Ribozyme Domain: Principles of RNA Packing. Science 273: 1678-1685
- ^ Toor N, Keating KS, Taylor SD, Pyle AM (2008) Science 320(5872):77-82.
- ^ Noller, H. (2005) RNA Structure: Reading the Ribosome. Science 308: 1508-1514
- ^ Lee, AJ. and Crothers, DM. The solution structure of an RNA loop-loop complex: the ColE1 inverted loop sequence. Structure (1998) 6, 993
- ^ Ferré-D'Amaré1, AR. Zhou, K. Doudna, JA. Crystal structure of a hepatitis delta virus ribozyme. Nature (1998) 395, 567-574
- ^ Celander DW, Cech TR (1991) Science 251:401–407.
- ^ Pyle, AM (2002) Metal Ions in the Structure and Function of RNA. J Biol Inorg Chem.
- ^ Cate, JH (1996) Metal-Binding sites in the major groove of a large ribozyme domain. Structure 4:10.
- ^ Kieft, JS (1997) Solution structure of a metal-binding site in the major groove of RNA complexed with cobalt(III)hexammine. Structure 5:713-21.
- ^ Rudisser, S (2000) Solution structure of cobalt(III)hexammine complexed to the GAAA tetraloop, and metal-ion binding to G·A mismatches. J Mol Bio 295:1211-23.
- ^ Burkhardt, C (2001) Modelling ion binding to AA platform motifs in RNA: a continuum solvent study including conformational adaptation. Nucleic Acids Research 21:19.
- ^ P. B. Moore 1999. Structural Motifs in RNA. Annual Reviews in Biochemistry. 68 :287-300
- ^ Abramovitz, D.L., Pyle, A.M. Remarkable morphological variability of a common RNA folding motif: the GNRA Tetraloop-receptor interaction. (1997) J. Mol. Biology, 266, 493-506
- ^ Moody, E.M., Feerrar, J.C., Bevilacqua, P.C. (2004) Evidence that Folding of a RNA Tetraloop Hairpin Is Less Cooperative that Its DNA Counterpart. Biochemistry, 43, 7002-7998
- ^ Blackburn et al. RNA Structure and Function: RNA Tertiary Structure, Nucleic Acids in Chemistry and Biology, Royal Society of Chemistry, 2006
- ^ Jaeger, L., Michel, F., Westhof, E. (1994) Involvement of A GNRA Tetraloop in Long-range RNA Tertiary Interaction, J. Molecular Biology.236, 1271-1276
- ^ Cate.J.H. et al. (1996) Crystal structure of a group I ribozyme domain: principles of RNA packing. Science, 273, 1678-1685
- ^ Michel F, Westhof E. J Mol Biol. 1990 Dec 5;216(3):585-610
- ^ Jaeger, L., Michel, F., Westhof, E. (1994) Involvement of A GNRA Tetraloop in Long-range RNA Tertiary Interaction, J. Molecular Biology, 236, 1271-1276
- ^ Cate.J.H. et al. (1996) Crystal structure of a group I ribozyme domain: principles of RNA packing. Science, 273, 1678-1685
- ^ Doherty EA et al., A universal mode of helix packing in RNA, Nature Structural Biology 8, 339 - 343 (2001)
- ^ Tom Steitz. RNA tertiary interactions in the large ribosomal subunit: the A-minor motif. PNAS, 2001
- ^ Pyle. Crystal structure of a self-spliced group II intron. Science, 2008
- ^ Satoko Yoshizawa, et al.Recognition of the Codon-Anticodon Helix Ribosomal RNA Science(1999) 285,1722
- ^ Batey, R.T., Rambo, R.P., Doudna, J.A. Tertiary Motifs in RNA Structure and Folding. Angewandte Chemie International Edition. (1999) 38, 2326.
- ^ Toor N., Keating, K.S., Fedorova, O., Rajashankar, K., Wang, J., Pyle, A.M. Tertiary architecture of the Oceanobacillus iheyensis group II intron. RNA (2010), 16, 57-69
- ^ Nature Structural Biology 8, 339 - 343 (2001)
- ^ Nature Structural Biology 5, 1037 - 1042 (1998)
- ^ Nature. 2000 Jul 20;406(6793):315-8.
- ^ Cheong C and Moore PB. (1992) Biochemistry 36:8406–8414.
- ^ Toor N, Keating KS, Taylor SD, Pyle AM (2008) Science 320(5872):77-82.
- ^ Feigon, J et al. (2008) J Mol Biol. 384(5): 1249-61.
- ^ Gilbert S, Rambo R, Van Tyne D, Batey R. (2008) Nat Struct Mol Biol. 15(2): 177-82.
- ^ Cheong, C. & Moore, P. B. (1992) Biochemistry 36, 8406–8414.
- ^ Robert T. Batey, etc. NATURE |VOL 432 | 18 NOVEMBER 2004
- ^ Baugh C, Grate D, Wilson C. J Mol Biol. 2000 Aug 4;301(1):117-28.
- ^ Arthanari H, Bolton PH. (2001) Chem. Biol. 8, 221-230
- ^ Robert T. Batey, etc. NATURE |VOL 432 | 18 NOVEMBER 2004
- ^ Oliver AW, et al. (2000) J. Mol. Biol.