Streamlining theory
Genomic streamlining is a theory in evolutionary biology and microbial ecology that suggests that there is a reproductive benefit to prokaryotes having a smaller genome size with less non-coding DNA and fewer non-essential genes.[1][2] There is a lot of variation in prokaryotic genome size, with the smallest free-living cell's genome being roughly ten times smaller than the largest prokaryote.[3] Two of the free-living bacterial taxa with the smallest genomes are Prochlorococcus and Pelagibacter ubique,[4][5] both highly abundant marine bacteria commonly found in oligotrophic regions. Similar reduced genomes have been found in uncultured marine bacteria, suggesting that genomic streamlining is a common feature of bacterioplankton.[6] This theory is typically used with reference to free-living organisms in oligotrophic environments.[1]
Overview
[edit]
Genome streamlining theory states that certain prokaryotic genomes tend to be small in size in comparison to other prokaryotes, and all eukaryotes, due to selection against the retention of non-coding DNA.[2][1] The known advantages of small genome size include faster genome replication for cell division, fewer nutrient requirements, and easier co-regulation of multiple related genes, because gene density typically increases with decreased genome size.[2] This means that an organism with a smaller genome is likely to be more successful, or have higher fitness, than one hindered by excessive amounts of unnecessary DNA, leading to selection for smaller genome sizes.[2]
Some mechanisms that are thought to underlie genome streamlining include deletion bias and purifying selection.[1] Deletion bias is the phenomenon in bacterial genomes where the rate of DNA loss is naturally higher than the rate of DNA acquisition.[2][7] This is a passive process that simply results from the difference in these two rates.[7] Purifying selection is the process by which extraneous genes are selected against, making organisms lacking this genetic material more successful by effectively reducing their genome size.[2][8] Genes and non-coding DNA segments that are less crucial for an organism survival will be more likely to be lost over time.[8]
This selective pressure is stronger in large marine prokaryotic populations, because intra-species competition favours fast, efficient and inexpensive replication.[2] This is because large population sizes increase competition among members of the same species, and thus increases selective pressure and causes the reduction in genome size to occur more readily among organisms of large population sizes, like bacteria.[2] This may explain why genome streamlining seems to be particularly prevalent in prokaryotic organisms, as they tend to have larger population sizes than eukaryotes.[9]
It has also been proposed that having a smaller genome can help minimize overall cell size, which increases a prokaryotes surface-area to volume ratio.[10] A higher surface-area to volume ratio allows for more nutrient uptake proportional to their size, which allows them to outcompete other larger organisms for nutrients.[11][10] This phenomenon has been noted particularly in nutrient depleted waters.[10]
Genomic signatures
[edit]Genomic analysis of streamlined organisms have shown that low GC content, low percentage of non-coding DNA, and a low fraction of genes encoding for cytoplasmic membrane proteins, periplasmic proteins, transcriptionally related proteins, and signal transduction pathways are all characteristic of free-living streamlined prokaryotic organisms.[6][4][12] Oftentimes, highly streamlined organisms are difficult to isolate by culturing in a laboratory (SAR11 being a central example).[6][4]
Model organisms
[edit]Pelagibacter ubique (SAR11)
[edit]Pelagibacter ubique are members of the SAR11 clade, a heterotrophic marine group which are found throughout the oceans and are rather common.[4] These microbes have the smallest genome and encode the smallest number of Open Reading Frames of any known non-sessile microorganism.[4] P. ubique has complete biosynthetic pathways and all necessary enzymes for the synthesis of 20 amino acids and only lack a few cofactors despite the genome's small size. The genome size for this microorganism is achieved by lack of, "pseudogenes, introns, transposons, extrachromosomal elements, or inteins". The genome also contains fewer paralogs compared to other members of the same clade and the shortest intergenic spacers for any living cell.[4] In these organisms, unusual nutrient requirements were found due to the streamlining selection and gene loss when selection occurred for more efficient resource utilization in oceans with limited nutrients for uptake.[13] These observations indicate that some microbes may be difficult to grow in a laboratory setting because of unusual nutrient requirements.[13]
Prochlorococcus
[edit]Prochlorococcus is one of the dominant cyanobacteria and is a main participant in primary production in oligotrophic waters.[14] It is the smallest and most abundant photosynthetic organism recorded on Earth.[14] As a cyanobacteria, they have an incredible ability to adapt to environments with very poor nutrient availability, as they maintain their energy from light.[15] The nitrogen assimilation pathway in this organism has been significantly modified to adapt to the nutritional limitations of the organisms’ habitats.[15] These adaptations led to the removal of key enzymes from the genome, such as nitrate reductase, nitrite reductase, and often urease.[15] Unlike some cyanobacterial counterparts, Prochlorococcus is not able to fix atmospheric nitrogen (N2).[16] The only nitrogen sources found to be used by this species are ammonia, which is incorporated into glutamate via the enzyme glutamine synthetase and uses less energy compared to nitrate usage, and in certain species, urea.[16] Moreover, metabolic regulation systems of Prochlorococcus were found to be greatly simplified.[15]
Nitrogen-fixing marine cyanobacteria (UCYN-A)
[edit]
Nitrogen-fixing marine cyanobacteria are known to support oxygen production in oceans by fixing inorganic nitrogen using the enzyme nitrogenase.[17] A special subset of these bacteria, UCYN-A, was found to lack the photosystem II complex usually used in photosynthesis and that it lacks a number of major metabolic pathways but is still capable of using the electron transport chain to generate energy from a light source.[17] Furthermore, anabolic enzymes needed for creating amino acids such as valine, leucine and isoleucine are missing, as well as some which lead to phenylalanine, tyrosine and tryptophan biosynthesis.
This organism seems to be an obligate photoheterotroph that uses carbon substrates for energy production and some biosynthetic materials for biosynthesis. It was discovered that UCYN-A developed a reduced genome of only 1.44 Megabases that is smaller but similar in structure to that of chloroplasts.[17] In comparison with related species such as Crocosphaera watsonii and Cyanothece sp., which employ genomes which range in length from 5.46 to 6.24 megabases, the UCYN-A genome is much smaller. The compacted genome is a single, circular chromosome with “1,214 identified protein-coding regions”.[17] The genome of UCYN-A is also highly conserved ( >97% nucleotide identity) across ocean waters, which is atypical of ocean microbes. The lack of UCYN-A genome diversity, presence of nitrogenase and hydrogenase enzymes for the TCA cycle, reduced genome size and coding efficiency of the DNA suggest that this microorganism may have symbiotic lifestyle and live in close association with a host. However, the true lifestyle of this microbe remains unknown.[17]
Alternative cases of small genomes
[edit]Bacterial symbionts, commensals, parasites, and pathogens
[edit]Bacterial symbionts, commensals, parasites, and pathogens often have even smaller genomes and fewer genes than free-living organisms, and non-pathogenic bacteria.[1] They reduce their "core" metabolic repertoire, making them more dependent on their host and environment.[1] Their genome reduction occurs by different evolutionary mechanisms than those of streamlined free-living organisms.[18] Pathogenic organisms are thought to undergo genome reduction due to genetic drift, rather than purifying selection.[18][1] Genetic drift is caused by small and effective populations within a microbial community, rather than large and dominating populations.[1] In this case, DNA mutations happen by chance, and thus often lead to maladaptive genome degradation and lower overall fitness.[18] Rather than losing non-coding DNA regions or extraneous genes to increase fitness during replication, they lose certain "core" metabolic genes that may now be supplemented by their host, symbiont, or environment.[18] Since their genome reduction is less dependent on fitness, pseudogenes are frequent in these organisms.[1] They also typically undergo low rates of horizontal gene transfer (HGT).
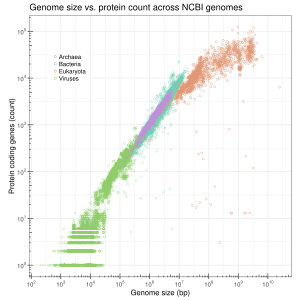
Viruses
[edit]Viral genomes resemble prokaryotic genomes in that they have very few non-coding regions.[19] They are, however, significantly smaller than prokaryotic genomes. While viruses are obligate intracellular parasites, viral genomes are considered streamlined due to the strong purifying selection that occurs when the virus has successfully infected a host.[20][21] During the initial phase of an infection, there is a large bottleneck for the virus population which allows for more genetic diversity, but due to the rapid replication of these viruses, the population size is restored quickly and the diversity within the population is reduced.[21]
RNA viruses in particular are known to have exceptionally small genomes.[22] This is at least in part due to the fact that they have overlapping genes.[22] By reducing their genome size, they increase their fitness due to faster replication.[22] The virus will then be able to increase population size more rapidly with faster replication rates.
Eukaryotes - birds
[edit]Genomic streamlining has been used to explain certain eukaryotic genome sizes as well, particularly bird genomes. Larger genomes require a larger nucleus, which typically translates to a larger cell size.[23] For this reason, many bird genomes have also been under selective pressure to decrease in size.[23][24] Flying with a larger mass due to larger cells is more energetically expensive than with a smaller mass.[24]
References
[edit]- ^ a b c d e f g h i Giovannoni SJ, Cameron Thrash J, Temperton B (August 2014). "Implications of streamlining theory for microbial ecology". The ISME Journal. 8 (8): 1553–65. Bibcode:2014ISMEJ...8.1553G. doi:10.1038/ismej.2014.60. PMC 4817614. PMID 24739623.
- ^ a b c d e f g h Sela I, Wolf YI, Koonin EV (October 2016). "Theory of prokaryotic genome evolution". Proceedings of the National Academy of Sciences of the United States of America. 113 (41): 11399–11407. Bibcode:2016PNAS..11311399S. doi:10.1073/pnas.1614083113. PMC 5068321. PMID 27702904.
- ^ Koonin EV, Wolf YI (December 2008). "Genomics of bacteria and archaea: the emerging dynamic view of the prokaryotic world". Nucleic Acids Research. 36 (21): 6688–719. doi:10.1093/nar/gkn668. PMC 2588523. PMID 18948295.
- ^ a b c d e f Giovannoni SJ, Tripp HJ, Givan S, Podar M, Vergin KL, Baptista D, Bibbs L, Eads J, Richardson TH, Noordewier M, Rappé MS, Short JM, Carrington JC, Mathur EJ (August 2005). "Genome streamlining in a cosmopolitan oceanic bacterium". Science. 309 (5738): 1242–5. Bibcode:2005Sci...309.1242G. doi:10.1126/science.1114057. PMID 16109880. S2CID 16221415.
- ^ Dufresne A, Salanoubat M, Partensky F, Artiguenave F, Axmann IM, Barbe V, Duprat S, Galperin MY, Koonin EV, Le Gall F, Makarova KS, Ostrowski M, Oztas S, Robert C, Rogozin IB, Scanlan DJ, Tandeau de Marsac N, Weissenbach J, Wincker P, Wolf YI, Hess WR (August 2003). "Genome sequence of the cyanobacterium Prochlorococcus marinus SS120, a nearly minimal oxyphototrophic genome". Proceedings of the National Academy of Sciences of the United States of America. 100 (17): 10020–5. doi:10.1073/pnas.1733211100. PMC 187748. PMID 12917486.
- ^ a b c Swan BK, Tupper B, Sczyrba A, Lauro FM, Martinez-Garcia M, González JM, Luo H, Wright JJ, Landry ZC, Hanson NW, Thompson BP, Poulton NJ, Schwientek P, Acinas SG, Giovannoni SJ, Moran MA, Hallam SJ, Cavicchioli R, Woyke T, Stepanauskas R (July 2013). "Prevalent genome streamlining and latitudinal divergence of planktonic bacteria in the surface ocean" (PDF). Proceedings of the National Academy of Sciences of the United States of America. 110 (28): 11463–8. Bibcode:2013PNAS..11011463S. doi:10.1073/pnas.1304246110. PMC 3710821. PMID 23801761.
- ^ a b Mira A, Ochman H, Moran NA (October 2001). "Deletional bias and the evolution of bacterial genomes". Trends in Genetics. 17 (10): 589–96. doi:10.1016/s0168-9525(01)02447-7. PMID 11585665.
- ^ a b Molina N, van Nimwegen E (January 2008). "Universal patterns of purifying selection at noncoding positions in bacteria". Genome Research. 18 (1): 148–60. doi:10.1101/gr.6759507. PMC 2134783. PMID 18032729.
- ^ Lynch M, Conery JS (November 2003). "The origins of genome complexity". Science. 302 (5649): 1401–4. Bibcode:2003Sci...302.1401L. CiteSeerX 10.1.1.135.974. doi:10.1126/science.1089370. PMID 14631042. S2CID 11246091.
- ^ a b c Chen B, Liu H (March 2010). "Relationships between phytoplankton growth and cell size in surface oceans: Interactive effects of temperature, nutrients, and grazing". Limnology and Oceanography. 55 (3): 965–972. Bibcode:2010LimOc..55..965C. doi:10.4319/lo.2010.55.3.0965.
- ^ Cotner JB, Biddanda BA (March 2002). "Small Players, Large Role: Microbial Influence on Biogeochemical Processes in Pelagic Aquatic Ecosystems". Ecosystems. 5 (2): 105–121. Bibcode:2002Ecosy...5..105C. CiteSeerX 10.1.1.484.7337. doi:10.1007/s10021-001-0059-3. S2CID 39074312.
- ^ Lauro FM, McDougald D, Thomas T, Williams TJ, Egan S, Rice S, DeMaere MZ, Ting L, Ertan H, Johnson J, Ferriera S, Lapidus A, Anderson I, Kyrpides N, Munk AC, Detter C, Han CS, Brown MV, Robb FT, Kjelleberg S, Cavicchioli R (September 2009). "The genomic basis of trophic strategy in marine bacteria". Proceedings of the National Academy of Sciences of the United States of America. 106 (37): 15527–33. Bibcode:2009PNAS..10615527L. doi:10.1073/pnas.0903507106. PMC 2739866. PMID 19805210.
- ^ a b Carini P, Steindler L, Beszteri S, Giovannoni SJ (March 2013). "Nutrient requirements for growth of the extreme oligotroph 'Candidatus Pelagibacter ubique' HTCC1062 on a defined medium". The ISME Journal. 7 (3): 592–602. Bibcode:2013ISMEJ...7..592C. doi:10.1038/ismej.2012.122. PMC 3578571. PMID 23096402.
- ^ a b Biller SJ, Berube PM, Lindell D, Chisholm SW (January 2015). "Prochlorococcus: the structure and function of collective diversity". Nature Reviews. Microbiology. 13 (1): 13–27. doi:10.1038/nrmicro3378. hdl:1721.1/97151. PMID 25435307. S2CID 18963108.
- ^ a b c d García-Fernández JM, de Marsac NT, Diez J (December 2004). "Streamlined regulation and gene loss as adaptive mechanisms in Prochlorococcus for optimized nitrogen utilization in oligotrophic environments". Microbiology and Molecular Biology Reviews. 68 (4): 630–8. doi:10.1128/MMBR.68.4.630-638.2004. PMC 539009. PMID 15590777.
- ^ a b Johnson ZI, Lin Y (June 2009). "Prochlorococcus: approved for export". Proceedings of the National Academy of Sciences of the United States of America. 106 (26): 10400–1. Bibcode:2009PNAS..10610400J. doi:10.1073/pnas.0905187106. PMC 2705537. PMID 19553202.
- ^ a b c d e Tripp HJ, Bench SR, Turk KA, Foster RA, Desany BA, Niazi F, Affourtit JP, Zehr JP (March 2010). "Metabolic streamlining in an open-ocean nitrogen-fixing cyanobacterium". Nature. 464 (7285): 90–4. Bibcode:2010Natur.464...90T. doi:10.1038/nature08786. PMID 20173737. S2CID 205219731.
- ^ a b c d Weinert LA, Welch JJ (December 2017). "Why Might Bacterial Pathogens Have Small Genomes?". Trends in Ecology & Evolution. 32 (12): 936–947. Bibcode:2017TEcoE..32..936W. doi:10.1016/j.tree.2017.09.006. PMID 29054300.
- ^ Koonin EV (February 2009). "Evolution of genome architecture". The International Journal of Biochemistry & Cell Biology. 41 (2): 298–306. doi:10.1016/j.biocel.2008.09.015. PMC 3272702. PMID 18929678.
- ^ Zwart MP, Erro E, van Oers MM, de Visser JA, Vlak JM (May 2008). "Low multiplicity of infection in vivo results in purifying selection against baculovirus deletion mutants". The Journal of General Virology. 89 (Pt 5): 1220–4. doi:10.1099/vir.0.83645-0. PMID 18420800.
- ^ a b Zwart MP, Willemsen A, Daròs JA, Elena SF (January 2014). "Experimental evolution of pseudogenization and gene loss in a plant RNA virus". Molecular Biology and Evolution. 31 (1): 121–34. doi:10.1093/molbev/mst175. hdl:10251/72658. PMC 3879446. PMID 24109604.
- ^ a b c Belshaw R, Pybus OG, Rambaut A (October 2007). "The evolution of genome compression and genomic novelty in RNA viruses". Genome Research. 17 (10): 1496–504. doi:10.1101/gr.6305707. PMC 1987338. PMID 17785537.
- ^ a b Kapusta A, Suh A, Feschotte C (February 2017). "Dynamics of genome size evolution in birds and mammals". Proceedings of the National Academy of Sciences of the United States of America. 114 (8): E1460–E1469. Bibcode:2017PNAS..114E1460K. doi:10.1073/pnas.1616702114. PMC 5338432. PMID 28179571.
- ^ a b Wright NA, Gregory TR, Witt CC (March 2014). "Metabolic 'engines' of flight drive genome size reduction in birds". Proceedings: Biological Sciences. 281 (1779): 20132780. doi:10.1098/rspb.2013.2780. PMC 3924074. PMID 24478299.
